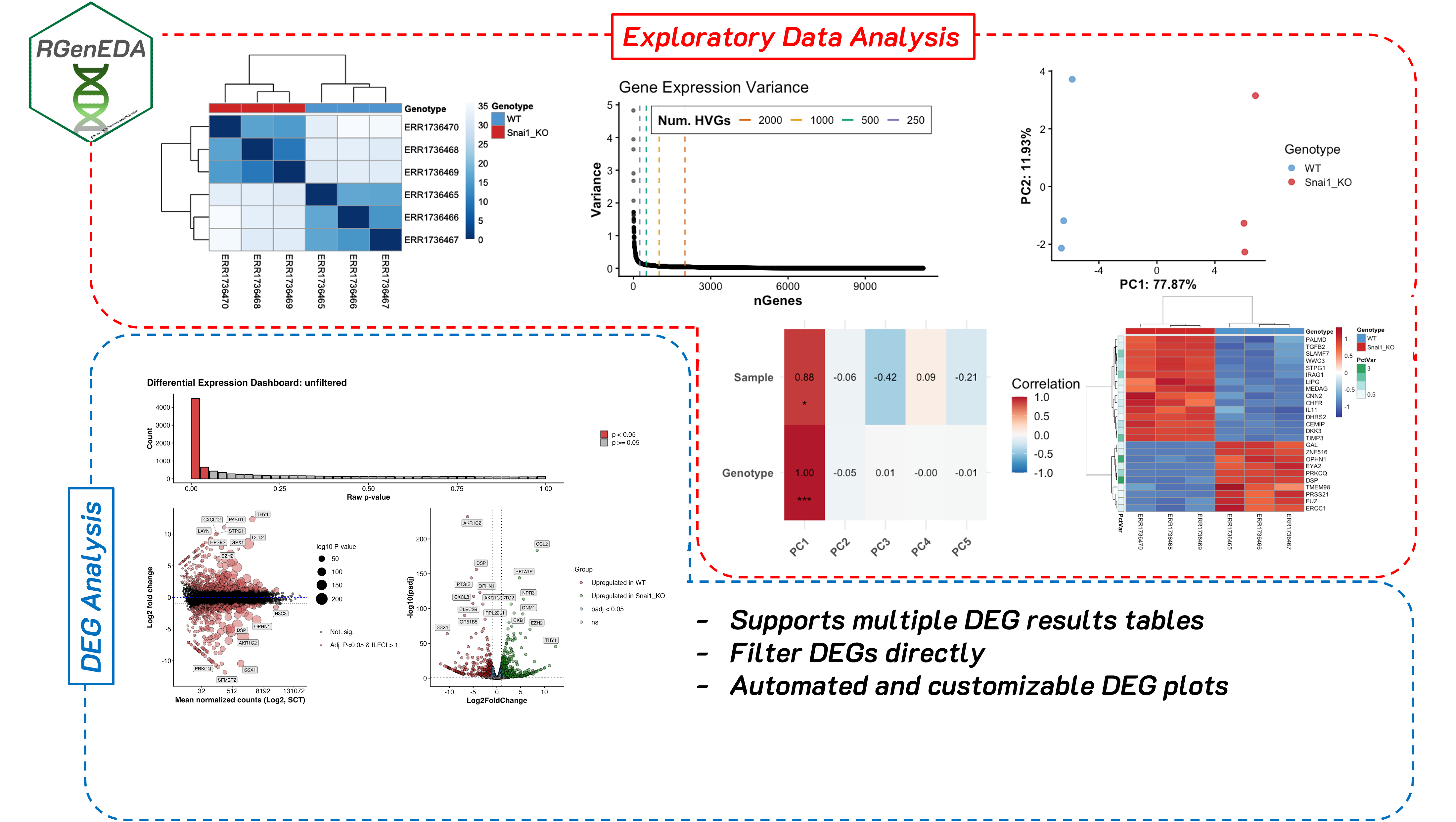
Clean, unified, streamlined, and reproducible frameworks for genomic exploratory data analyses
Development
[!IMPORTANT] RenEDA is currently under active development. If there is a feature you would like implemented, please submit a feature request on the Issues page of submit a pull request. For information on new additions see the Change Log
[!WARNING] RGenEDA has updated to v2.0.0 which introduced some breaking changes. Please ensure you are using the most up-to-date version
Installation
Install the latest version from Github
install.packages("devtools") # if not already instaled
library(devtools)
devtools::install_github("mikemartinez99/RGenEDA")
library(RGenEDA)Usage
For a full demo, see Snail1 KO Dataset Demo
Change-log


- Implemented core, standalone functions
- Variance calculation, PCA calculation, and Eucliden distances, and Eigenvector correlations


- Introduced S4 GenEDA object for streamlined data handling
- Added methods for HVG calculation and PCA storage within S4 slots
- Added visualization functions
- PCA plots
- Variance plots for HVGs
- Eigenvector (gene-loading) plots
- Revamped eigencorrelation plots to only use top number of HVGs identified in HVG selection/PCA
- Enhanced plot output flexibility (return ggplot2, pheatmap, or ComplexHeatmap objects for full customization
- Added unit tests
- Added Pasilla dataset vignette

November 3rd, 2025
- Rely soley on Pheatmap for all heatmap visualizations
- Fixed top-gene-loading bug for selected PCs in PlotEigenHeatmap function
- Added optional DEGs slot to GenEDA object to interface with DESeq2
- Added MA plot functionality

November 10th, 2025
- Fixed example blocks in roxygen headers
- Addressed all R cmd build errors, warnings, and notes

November 15th, 2025: “The DEG update”
- Added assay argument to SetDEGs function
- Added assay and saveAssay argument to FilterDEGs function to specify which assay to filter and what to save filtered assay as
- Added assay argument to PlotMA to specify which DEG slot to plot
- Added PlotVolcano function for direct volcano plotting functionality
- Added FindHVDEGs to intersect DEGs with HVGs calculated during EDA functions
- Added GenSave function to save save pheatmap objects in a manner similar to ggplpot2::ggsave()
- Renamed eigencorr, ordcorr, and distanceHeatmap functions to PlotEigenCorr, PlotOrdCorr, and PlotDistances respectively to match other plotting function name conventions
- Replaced Pasilla dataset demo with Snail1 KO dataset from Matsuri et al., 2018



